BIOCHEMISTRY TOPICS
Electron transport chain
The multisubunit membrane complexes of the electron transport chain (ETC). Prosthetic groups and mobile electron carriers. Coupling electron transport to proton translocation. Inhibitors of electron transport.
The electron transport chain (ETC)
The ETC is responsible for the reduction of molecular oxygen by NADH. This exergonic process (electrons from NADH enter at a relatively low E°′, and electrons exit at relatively high E°′ as they reduce O2 to H2O. making ΔE°′ positive, and thus ΔG°′ is negative) is carried out in a precisely controlled, multistep manner that preserves much of the energy released in the form of a transmembrane electrochemical gradient. This feat is accomplished by four integral membrane protein complexes,. NADH-Q oxidoreductase (Complex I) [EC 1.6.5.3], succinate-Q reductase (Complex II) [EC 1.3.5.1], quinol--cytochrome-c reductase (Complex III) [EC 1.10.2.2], and cytochrome c oxidase (Complex IV) [EC 1.9.3.1]. Each of the complexes moves electrons along a path within its structure formed by a series of fixed, redox-active prosthetic groups, spaced through the structure in precise intervals and orientations with respect to one another. These prosthetic groups include flavin nucleotides (FAD and FMN), quinones, iron-sulfur (Fe-S) clusters, heme, and copper ion. Two mobile electron carriers shuttle electrons between these complexes, coenzyme Q (ubiquinone) and cytochrome c. Three of the four complexes translocate protons across the inner mitochondrial membrane, thus generating the transmembrane electrochemical gradient. Five types of prosthetic groups are involved in electron transfer within the complexes.
Complex I - NADH-Q oxidoreductase (Complex I) [EC 1.6.5.3]
Official name: NADH:ubiquinone reductase (H+-translocating).
Substrates: (reductant) nicotinamide adenine dinucleotide (hydride, NADH); (oxidant) Coenzyme Q (Q, ubiquinone)
Products : nicotinamide adenine dinucleotide (NAD+); Coenzyme Q (QH2, ubiquinol)
Cofactors (prosthetic groups):
flavin mononucleotide (FMN), iron-sulfur (Fe-S) clusters.
Protons pumped: about 4 H+ per pair of electrons (= 1 NADH) transferred.
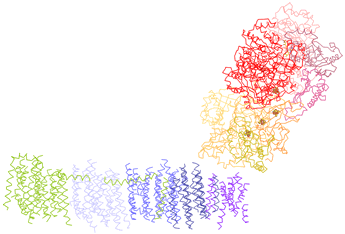
Complex I is the largest of the mitochondrial electron transport complexes (>900 kD in size) with 46 distinct polypeptide chains (a simpler bacterial version is shown in the figure at left). The structure can be described as contained within a roughly L-shaped (a lazy L, that is lying on its back) envelope, with a peripheral arm protruding into the matrix and a membrane-bound portion forming the other arm. In this figure - and those for the other ETC complexes below - the view is perpendicular to the plane of the membrane, which would be visualized as a plane extending out from and behind the screen. With respect to an inner mitochondrial membrane, the view is oriented such that the matrix (inner side) compartment lies above, and the intermembrane space (outer side) lies below. Here, for Complex I, the intersection of the membrane plane and the structure of the complex corresponds to the horizontal arm of the L, the membrane-bound portion. The peripheral arm contains an FMN prosthetic group that presumably acts as the direct electron acceptor from NADH, and a series of iron-sulfur clusters (visible in the figure within the yellow, orange, and split pea colored subunits in the figure in the lower part of the peripheral arm). A site for binding and reduction of coenzyme Q (quinone form) is near the vertex of the L. The proton pumping subunits comprise the membrane-bound arm of the L.
Complex II - Succinate dehydrogenase
Official name: Succinate dehydrogenase (quinone).
Alternate name: Succinate-coenzyme Q reductase (SQR).
Substrates: (reductant) succinate (citric acid cycle intermediate); (oxidant) Coenzyme Q (Q, ubiquinone)
Products : fumarate (citric acid cycle intermediate); Coenzyme Q (QH2, ubiquinol)
Cofactors (prosthetic groups):
flavin adenine dinucleotide (FAD), heme b, iron-sulfur (Fe-S) clusters, Coenzyme Q.
Protons pumped: none.
Complex II, which is the simplest of the ETC complexes, consists of four subunits (pictured at left).
Complex II forms a direct link to the citric acid cycle. A flavin adenine dinucleotide (FAD) prosthetic group accepts electrons from succinate, forming fumarate and FADH2. The latter transfers electrons through a series of iron-sulfur clusters, and are then conducted, with the assistance of a nearby heme, to an exchangeable quinone bound in a site in the "stalk" of the complex (the blue and green subunits in the figure), which is embedded within the membrane. Complex II catalyzes electron transfer, but unlike the other ETC complexes, it does not couple it to proton translocation.
Above, left: Ribbon diagram of the Complex II structure from E. coli.
Each of the four subunits of the mushroom-shaped assembly is shown in a different color. The two hydrophilic subunits (purple and orange)
project into the inner compartment (for bacteria such as E. coli, the cytosol; in mitochondria, the matrix),
and two membrane-spanning subunits (green and blue) make up a stalk.
The hydrophilic subunits appear stacked with the flavoprotein
(the locus of succinate dehydrogenase activity, possessing a covalently-linked FAD) in purple on top,
and an iron-sulfur protein (ISP, in orange) below it. The ISP has three different iron-sulfur clusters that line up providing
an electron transfer path (see figure at right) between FADH2 and a bound coenzyme Q in one of the integral membrane subunits.
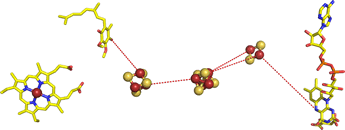
Above, right:The arrangement of prosthetic groups in the
succinate-Q reductase (Complex II) structure,
with the protein not displayed.
The view is a 90° clockwise rotation relative to the view seen in the ribbon diagram at left.
From left to right is heme,
a Q2 molecule (ubiquinone)
with its nonpolar isoprenoid tail shortened to two five-carbon units), three
iron-sulfur clusters, and FAD.
The red dashes are meant to suggest electron transfer paths through the protein.
Electrons move from right to left.
The iron-sulfur clusters are separated by about 9 and 11 Å. Both figures were drawn from pdb
1nek.
using PyMOL (see reference below).
Q-cytochrome c oxidoreductase (Complex III)
250 kD, 11 subunits. Cofactors: heme, Fe-S. Two H+ pumped per electron pair transferred. Also called cytochrome bc1 complex.
The subunits of the complex that are of principal interest are a cytochrome b protein (carrying two heme b prosthetic groups), a cytochrome c1 protein, and a Rieske iron-sulfur protein (ISP).
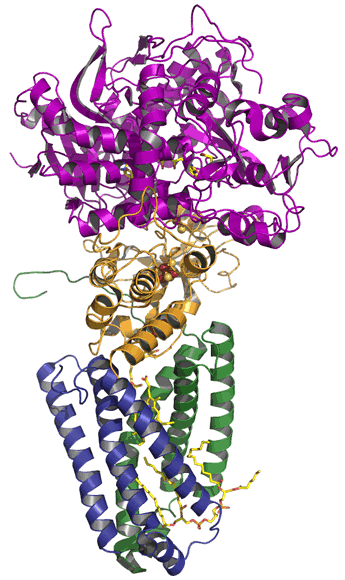
Left: The structure of Complex III (cytochrome bc1 complex) from chicken mitochondria, with the polypeptide chains represented as C-alpha traces. The view is perpendicular to the plane of the inner mitochondrial membrane, with the matrix above. An assembly of 10 distinct polypeptide chains form one half of the structure - these are represented in different colors - and a second set of the same 10 chains comprise the second half of the structure (shown in light blue). The prosthetic groups are visible, in part, as stick representations. In a counterclockwise arc from the bottom right to near the midpoint of the lower portion of the dimer are: a c-type heme, an iron-sulfur cluster (associated with the Rieske iron-sulfur protein, in sky blue), stigmatellin (occupying the outer Q site), and a bL heme group. The molecules above it are (left to right) coenzyme Q (occupying the inner Q site, a bH heme group, and cardiolipin. This figure was drawn from pdb 3h1j. using PyMOL (see reference below).
Electron transport through Complex III occurs by means of a two-stage redox loop called the Q cycle. The net result of the Q cycle is the transfer of the two electrons from a molecule of reduced coenzyme Q (ubiquinol or QH2) to two molecules of oxidized cytochrome c forming two molecules of reduced cytochrome c and a molecule of oxidized coenzyme Q (ubiquinone or Q). This redox reaction is accompanied by the vectorial transfer of protons from the mitochondrial matrix (denoted as the N side of the membrane, upper side with respect to the protein structure shown in the figure) to the intermembrane space (P side of the membrane).
Cytochrome c oxidase (Complex IV)
160 kD, 13 subunits. Cofactors: hemes, copper ions. About 4 H+ are pumped out of the matrix per 2e- transferred.
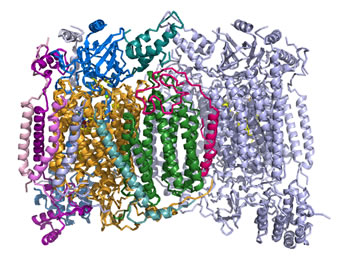
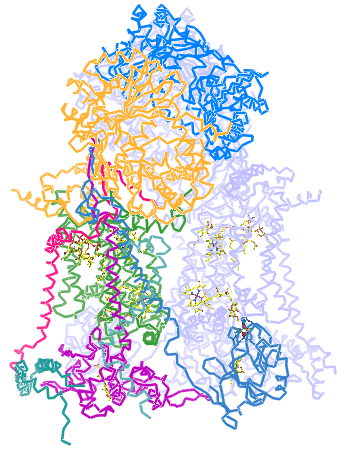
Right: Ribbon diagram of the cytochrome c oxidase (Complex IV) structure. Two copies of a multisubunit assembly of 13 separate polypeptide chains and their associated prosthetic groups come together as a dimer. One copy of the dimer is shown with the individual chains represented in different colors, with the other copy shown in light blue. The view is perpendicular to the plane of the inner mitochondrial membrane, with the matrix above. The figure was drawn from pdb 1occ. using PyMOL (see reference below).
The ETC in context: Oxidative phosphorylation
The figure below illustrates the coupling of catabolic metabolism to generation biochemical energy (ATP) in a summary cartoon. In the stepwise biochemical process of glucose oxidation to water and carbon dioxide, electrons are fed into the so-called electron transport chain. The three vertical ovals in this cartoon represent the membrane-embedded protein complexes that carry out the ETC (note that the orientation of the membrane is flipped relative to the structral figures above, with matrix below and intermembrane space above) which couples electron flow, which to vectorial movement of hydrogen ions (Step 1, three upward arrows). This generates the free energy of an electrochemical gradient (specifically, a proton gradient, or a transmembrane ΔpH). The energy of this gradient is harvested when the H+ are allowed to move back across the membrane (Step 2) to drive the synthesis of ATP from ADP and phosphate (Step 3).
This is carried out in cells by a transmembrane protein complex called ATP synthase. In essence, the energy released by chemical reactions is transduced into a different form of potential energy associated with the physical separation of charged species across a membrane, a physicochemical form of energy. This energy is in turn transduced into the biochemical energy "currency", ATP.
In oxidative phosphorylation, the free energy of the transmembrane difference in pH, created by the oxidation of metabolic fuel to water and carbon dioxide, as taken to completion by the electron transport chain is in turn utilized to drive phosphorylation of ADP to produce ATP.
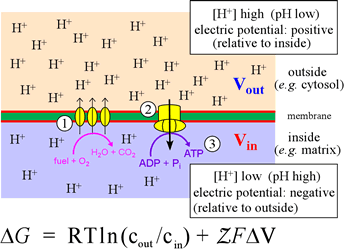
Left: Schematic summary of oxidative phosphorylation, illustrating the transmembrane pH difference and its relation to the electron transport chain and ATP synthesis. The equation is an expression for the Gibbs free energy of the electrochemical gradient. The term ΔV is defined as ΔV = Vout − Vin. The definition of pH can be used to write the first term of the equation as −2.3026RTΔpH (at 25° C).
Inhibitors of electron transport
Various inhibitors have been discovered, the use of which in research have illuminated the sequence of carriers and complexes shown in the scheme above. They have been used to show, for example, that there must be three distinct entry points for electrons to enter into the ETC and contribute to the generation of the proton gradient.
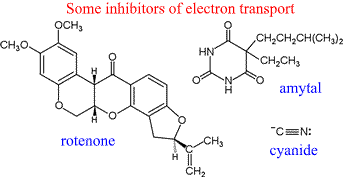
Inhibitors of electron transport through Complex I include rotenone and amytal. Rotenone is a plant-derived substance used by Amazonian tribes to poison fish. It has also been employed as an insecticide. Amytal is a type of barbiturate.
Electron transport through Complex III is inhibited by the antibiotic compound antimycin A.
Several small molecules and anions disrupt the reduction of oxygen in Complex IV by coordinating to the iron of heme a3. Cyanide and azide bind to the Fe(III) form of heme a3, while carbon monoxide binds its Fe(II) form. The latter is analogous to the strong affinity of CO for the heme iron in hemoglobin Recall that the functional form of iron in Hb is the +2 (ferrous) oxidation state, when it can bind oxygen or carbon monoxide.