Lecture 4. Nucleotides and nucleic acids
Friday 9 September 2016
Nucleotides: Nomenclature and chemical structure. Roles of nucleotides. Polynucleotides and nucleic acids. Tautomerism and base pairing; Chargaff’s rules. Structural features of the DNA double helix. Overview of nucleic acid function.
Reading: VVP4e - Ch.3, pp.40-51; Ch.24, pp.821-827.
Summary

Nucleic acids are one the major classes of biological macromolecules. The nucleic acids DNA and RNA are the carriers of information in living cells and viruses. DNA (deoxyribonucleic acid) is the physical realization of the genes and genomes of organisms, while RNA (ribonucleic acid) in most cases mediates expression of the genetic information stored in DNA. Nucleic acids are typically long polymers built up of repeating units called nucleotides. A nucleotide is a combination of a base with a sugar (usually ribose or deoxyribose) and phosphate. In the context of nucleotides, the bases are heterocyclic aromatic compounds of two general types: the purines, featuring a structure with two fused nitrogen-containing rings, and pyrimidines with a single ring. Nucleotides are classed specifically as ribonucleotides or deoxyribonucleotides (depending on the identity of the sugar component). Ribonucleotides are the monomeric units of RNA, while DNA is composed of deoxyribonucleotides. A nucleoside is just the base and sugar part of a nucleotide, making the latter a phosphate ester (or phosphoester) of a nucleoside.
The sugar component provides hydroxyl groups to participate in phosphoester linkages with the phosphate groups that occur in nucleotides. Free nucleotides most commonly have at least one phosphate attached to the hydroxyl of the exocyclic carbon 5 (this is denoted as the 5′position, where the prime indicates an atom of the sugar component).
The attachment of the nitrogen-containing bases is through an N-glycosidic bond to the anomeric C1′of the sugar component. We'll look more carefully at the glycosidic bonds when we study carbohydrates later in the course.
Nucleotides can be joined together in a series of phosphodiester linkages to form a nucleotide polymer, or polynucleotide. For example, a free 3' hydroxyl of one nucleotide can form a phosphoester linkage with a phosphate group attached to the 5' hydroxyl group of another to form a dinucleotide. The chain can be extended to arbitrary length by formation of additional phosphodiester bonds with more nucleotides. This forms a sugar-phosphate backbone. There is a directionality to the chain that is specified notationally as the 5'→3' or 3'→5' directions, by reference to the 5' position (hydroxymethylene group exo to the aldopentose ring of ribose or deoxyribose) and the 3' position (3' OH attached directly to the ring) of the sugar part of the backbone.
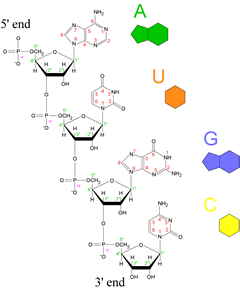
Left: Structural drawing of a polyribonucleotide, along with symbolic representations of the four common bases of ribonucleotides and RNA molecules to the right of the chemical structure. The directionality of the chain is indicated, with the 5' end at the top and the 3' end at the bottom.
By convention, a polynucleotide sequence can be represented by the single letter symbol for a base, with the first letter listed the 5' end and the last letter listed the 3' end. The four common ribonucleotides of RNA - along with their base components - are adenylate, which contains the base adenine (A); uridylate, which contains uracil (U), guanylate , containing guanine (G), and cytidylate which has the base cytosine (C).
Base pairing
Base pairing provides the chemical complementarity that underlies replication. In RNA sequences, G pairs with C and A pairs with U. Thus, the sequence in the figure would be written as AUGC and its reverse complement (for base-pairing in a double-stranded structure) is GCAU. (Note: In a DNA sequence, T replaces U.)
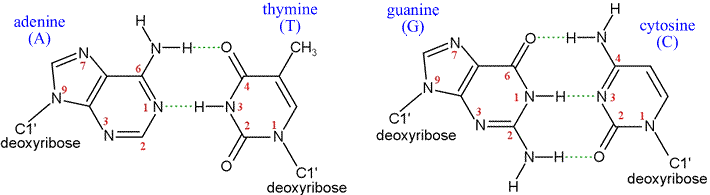
Above: Structural formulas for the two canonical Watson-Crick base pairs. Below, left: Examples of experimentally-observed base-pair strucures. The model is shown in stick form, where the sticks represent bonds, and atoms are centered at the ends or junctions of the sticks. The color code for atom type is as follows: carbon - yellow, nitrogen - blue, oxygen - red, phosphorous - pink. Hydrogen atoms are not shown - their locations are implied. The dashes indicate hydrogen bonds. Note the 2'-endo conformation of the deoxyribose rings, as well as the anti- orientation of the bases relative to the sugars about the N-glycosidic bonds. 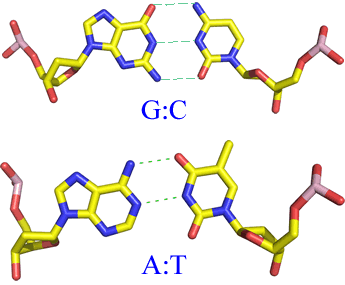
Double-stranded DNA
When two polynucleotide sequences show reverse complementarity, they can form an extended stretch of a perfectly base-paired double-stranded structure in which two single-stranded molecules align in opposite directions (the strands are said to be antiparallel), with their register specified by the many hydrogen bonds. DNA is usually found in vivo to be in such a double-stranded form, The chemical nature of the bases dictate that the purine base A pairs with T (a pyrimidine), and that C (pyrimidine base) pairs with G (purine base). This specificity in base pairing is the chemical basis of the conservation and duplication of genetic information. The base sequence of a single strand determines the sequence of its paired, or complementary strand. The stereochemical form of double-stranded DNA is a double helix, in which the sugar-phosphate backbones of both strands adopt a helical conformation that wrap around the outside of an imaginary cylinder, with the bases of each strand projecting toward each other on the inside, properly spaced and oriented for hydrogen bond-mediated pairing. The elucidation of this double helical structure of DNA by Watson and Crick in 1953 immediately suggested the chemical basis of heredity, laid the foundation for the science of molecular biology, and still remains the classic example of the way in which biological function follows from the structures of the relevant molecules.
The double helical structure of DNA can take several forms. The most common form, that discovered by Watson and Crick, is known as B form DNA, and its regular form is charaterized by right-handed helices, base pairs nearly perpendicular and close to the central axis of the helix, 10 base pairs per turn (36° twist), a spacing of 3.4 Å between base pairs (helix pitch of 34 Å), and a C2'-endo conformation of the deoxyribose sugar rings. The A form geometry, which is characteristic of double-stranded RNA, differs in the inclination and displacement of the base pairs with respect to the helix axis, and shows 11 base pairs per turn (32.7° twist), a 2.9 Å rise per base pair (helix pitch of 32 Å), and a C3'-endo conformation of the sugar. A third double helical form, Z DNA, is relatively rare. It is a left handed helix and its occurrence is limited to DNA sequences in which purine and pyrimidine bases alternate, mainly GC.
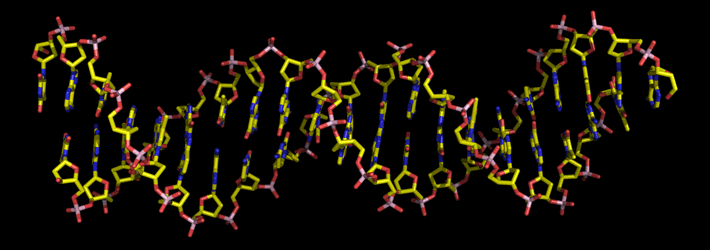
Above: A 20-base pair stretch of a DNA double helix. The stick model shows chemical bonds, with ends and junctions of the sticks representing the atom positions. Carbon is shown in yellow, oxygen in red, nitrogen blue, and phosphorous pink. Hydrogen atoms are not shown. The helix axis runs horizontally, and the base pairs are viewed nearly edge-on. The phosphate groups and deoxyribose rings of the antiparallel backbones are evident.
Diversity in the RNA world
RNA molecules show great diversity in structure and function. A single-stranded polyribonuclotide can fold back upon itself to form a variety of hairpin and stem-loop structures containing antiparallel, double-stranded helical regions featuring the canonical, Watson-Crick base pairs shown above. The favored confomation for an RNA helix is the A-form. This pattern of base-paired and loop regions can be thought of as RNA secondary structure (by analogy to protein structural hierarchy).
Untranslated RNA species are now being implicated in disease. Mutant forms of these RNAs have recently been linked to Fragile X tremor / ataxia syndrome (FXTAS) and myotonic dystrophy.
A ribozyme is an RNA molecule that catalyzes a chemical reaction. Examples include the self-splicing Group I intron from Tetrahymena, the "hammerhead" ribozyme, and the hairpin ribozyme. The RNA components of the ribosome could also be included, as biochemical, genetic, and structural evidence shows that the RNA, and not the protein components, catalyze the reactions of protein synthesis. The catalytic repertoire of ribozymes is limited mainly to phosphoryl transesterification and hydrolysis reactions.