Lecture 12. Enzyme kinetics and inhibition
Tuesday 26 February 2019
Enzyme kinetics and the Michaelis-Menten equation. Kinetic parameters Vmax and KM. Models for bisubstrate reactions.
Reading: Lehninger - Ch.6, pp.198-213.
Summary
Reading summary. §6.3
Enzyme kinetics as an approach to understanding mechanism.
(bottom, p.206f) Enzymes are subject to reversible or irreversible inhibition.
Box 6-2: Kinetic tests for determining inhibition mechanisms (p.209).
Table 6-9: Effects of reversible inhibitors on apparent Vmax and apparent KM
Box 6-3: -Medicine- Curing African sleeping sickness with a biochemical Trojan horse (p.211).
***
Enzyme kinetics catalytic principles, reaction coordinate diagrams, mechanistic models, rate constants and kinetic parameters.
The Michaelis-Menten equation describes a hyperbolic initial rate vs substrate concentration curve. Two parameters, accessible experimentally, KM and Vmax describe the hyperbolic curve.
We introduce the topic of enzyme inhibition by considering the mechanistic model for the phenomenon of competitive inhibition. Enzyme inhibitors are not only important tools for enzymological research, but also more broadly significant in biochemistry, biology, and medicine. Many drugs, for example, are specific inhibitors of particular enzymes. Aspirin is an irreversible inhibitor of cyclooxygenase activity.
Enzyme inhibition
The specificity of enzymes is not strictly limited to substrates. Often, the activity of an enzyme is reduced by specific interactions with molecules termed inhibitors. Enzyme inhibition is one of the most important phenomena in biochemistry. For example, many drugs, antibiotics, and toxins exert their effects by ther ability to inhibit an enzyme. Inhibitors that are structurally similar to the substrate are often competitive inhibitors, since they compete for binding at the active site. Enzyme inhibiton can be reversible (as is usually the case when an inhibitor binds to the enzyme via noncovalent interactions) or irreversible (as occurs in numerous cases where inhibitors act via covalent modifications to the enzyme, perhaps targeting a critical residue for catalysis).
We can imagine several simple models for reversible inhibition. The simplest of these is the direct occlusion of the active site by the inhibitor. This would be seen in the case of a molecule with some structural similarity to substrate. Binding of substrate and inhibitor are mutually exclusive in this model for competitive inhibition.
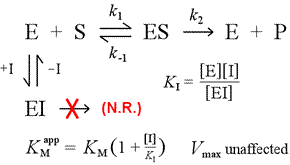
At right is shown a simple mechanistic model for competitive inhibition. The inhibitor, I, binds only to the free enzyme E, with a dissociation constant KI , and blocks substrate (S) binding. By tying up some of the enzyme in the inactive EI complex, less of it is available at a given substrate concentration to combine with substrate and form ES and then potentially convert to products. An increase in [S] necessary to reach ½Vmax will be observed, hence the apparent value of KM increases.
By increasing [S] to ever greater levels, the substrate can overwhelm the inhibitor, outcompeting it for the free enzyme to the extent that the true Vmax can still be approached.
We can easily imagine that a molecule that resembles the substrate in certain key structural features could compete with the substrate for binding the at the active site. This is called a substrate analog, and they provide common examples of competitive inhibitors of enzymes. If the way enzymes enhance reaction rates [once the enzyme binds with specificity its substrate(s)] is by selectively stabilizing the transition state of the reaction, then if we can find a molecule that resembles the transition state (in structure and polarity or charge), such a transition state analog ought to be a very good competitive inhibitor.
We'll contrast the competitive inhibition model with uncompetitive inhibition, in which the inhibitor binds only to the enzyme-substrate complex. One can imagine this occuring as a result of an induced-fit type enzyme-substrate interaction, in which a binding site for an inhibitor is available exclusively in the induced conformation of ES.
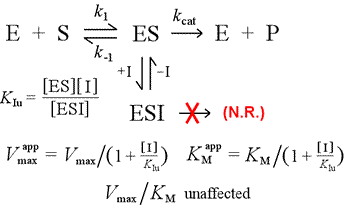
Left: A mechanistic model for uncompetitive inhibition. In this model, the inhibitor binds only the ES complex, and not free enzyme. The ternary complex, ESI, does not proceed to products. This has the effect of lowering the apparent Vmax. The inhibitor dissociation constant for ESI is denoted KIu. In an effect explained by Le Châtelier's Principle, the apparent KM is lower, as the inhibitor binding removes some of the product (ES) of the ES formation "equilibrium".
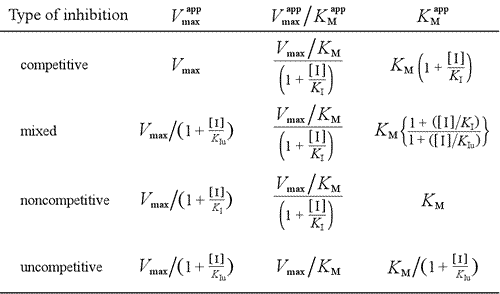
A model for inhibition in which inhibitor binds both free enzyme and the enzyme-substrate complex is mixed inhibition. The inhibitor dissociation constant may differ between E and ES (i.e. KI ≠ KIu). The special case where KI = KIu goes by the name noncompetitive inhibition. Note that in this case, KM is not affected, while Vmax is lowered.
Types of reversible enzyme inhibition - a summary
We have seen that the different models for reversible inhibition can be distinguished according to effects on kinetic parameters. The table below summarizes the types of inhibition and their effects on these parameters.
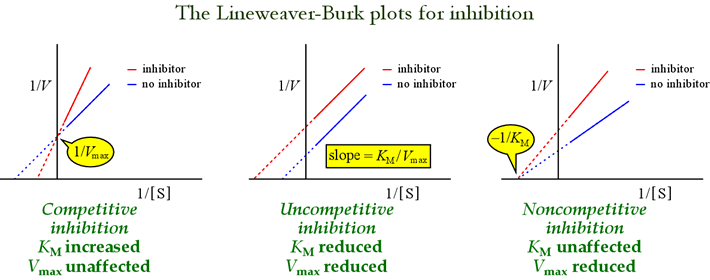
The Lineweaver-Burk, or double-reciprocal plots are useful for identifying patterns of inhibition. The figure below shows how different types of inhibition affect the plot.