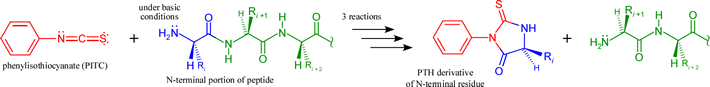Lecture 7. Protein primary structure
Monday 19 September 2016
Protein sequencing. Sequence databases and sequence comparisons - sequence alignments. Protein evolution: domains and molecular clocks.
Reading: VVP4e - Ch.5, pp.93-123.
Summary
Further exploration of protein chemistry and sequencing methods. Evolutionary implications of sequence comparisons.
Protein sequencing
Most proteins are much too large to sequence directly. Instead, a "divide and conquer" strategy is used, by which smaller peptide fragments are produced, followed by sequencing of the fragments by Edman degradation or mass spectrometry. If at least two distinct methods of fragmenting the protein are used, the assembly of the sequence is made tractable by the overlapping fragments produced. Two general methods are used to produce peptide fragments: endopeptidases, or proteases, which are enzymes that cleave the peptide bonds between amino acids within the polypeptide chain, and chemical methods.
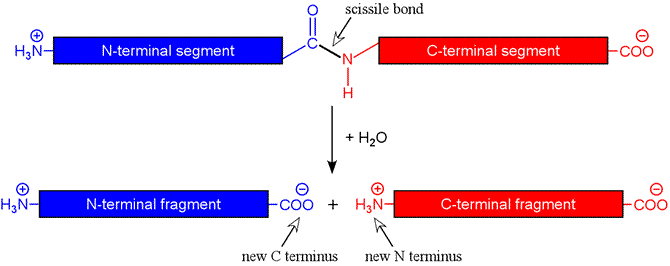
The figure below shows schematically the hydrolysis reaction catalyzed by a protease. The peptide bond targeted by a protease is termed the scissile bond. For an endopeptidase, the scissile bond is internal to the chain, i.e. the protease is is not acting on the first (or N-terminal) or last (C-terminal) peptide bonds. In the latter case, the term exopeptidase is used, and there are exopeptidases specific for one end or the other (aminopeptidases and carboxypeptidases).
For the approach to protein sequencing we are considering, endopeptidases are most useful. Furthermore, it is helpful if the endopeptidase used is specific - that is, it cleaves peptide bonds not in a random fashion, but only at certain locations in the chain, such as on the C-terminal side of certain amino acids. Ideally, the fragments yielded by treatment of a given protein with an endopeptidase are generated predictably and reproducibly. Fortunately, a degree of specificity is characteristic of enzymes in general. In fact, it is possible for the enzyme to be too specific in this context. A proteolytic enzyme employed for protein sequencing ideally acts with a moderate degree of specificity so that treatment of a protein with the enzyme yields an appropriate number and length of peptide fragments.
Two digestive enzymes of the serine protease family are useful in this sense, trypsin and chymotrypsin. Trypsin cleaves on the C-terminal side of Arg or Lys residues, although not if the following residue is Pro. Chymotrypsin is somewhat more loosely specific, cleaving on the C-terminal side of residues with large nonpolar sidechains, preferentially acting on the peptide bond following Phe, Tyr, and Trp (although again, not if the next residue is Pro).
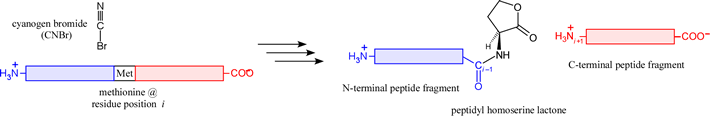
There are a few chemical methods with a specificity making them suitable for the generation of peptide fragments. The prime example is cyanogen bromide (CNBr), a reagent that cleaves on the C-terminal side of Met residues, generating a peptidyl homoserine lactone N-terminal fragment.
Edman degradation
Peptides generated by the above described (or similar) methods can in most cases be readily sequenced by the iterative chemical procedure known as Edman degradation. The procedure is based on a three-stage reaction that labels and removes the N-terminal residue of a polypeptide, which can be identified as a PTH (phenylthiohydantoin) derivative. Thus, the product peptide following the three steps is again a peptide that is one residue shorter at its N-terminus. This product can be then be subjected to another round of the same reactions. Below the overall reactants and products of one round of the three-step Edman procedure are shown, Note that the initial reaction requires the N-terminal amino group to be nucleophilic, hence the pH must be high enough to insure that this group is in its neutral base form.
In favorable cases, the process can be repeated many times, the liberated PTH-amino acid identified each time, and up to 100 residues of sequence determined in this manner. Furthermore, with technological advancement permitting automation and incorporating highly sensitive means of detection, Edman degradation can be performed on small amounts of a peptide - 5-10 pmol or <0.1 μg.
Sequencing by mass spectrometry
Mass spectrometry is an analytical method that measures mass-to-charge ratio (m/z) for ions in gas phase. Generally, the larger a molecules, the lower its vapor pressure. Hence, a major hurdle to application of mass spectrometry to biomolecules was to produce gas phase ions of an intact large polymer such as a protein or peptide. Work overcoming this challenge was rewarded by half of the The Nobel Prize in Chemistry 2002.
Electrospray ionization (ESI) is capable of producing multiply-charged intact polypeptides in gas phase. ESI mass spectrometry is an extremely useful and accurate method for determining mass of polypeptides and proteins. A family of ions is produced from a polypeptide that differ in both mass and charge by 1 atomic unit (based upon number of H+ taken up by proton accepting groups of the molecule). Our text (VVP4e) shows (Fig. 5-17, p.112) a mass spectrum of such a family for horse heart myoglobin, and demonstrates (Sample Calculation 5-1, p.113) how the m/z values for two successive peaks can used in an algebraic determination of the molecular mass.
Peptides of up to 25 residues can be sequenced by mass spectrometry. In tandem mass spectrometry (MS/MS), an analyte selected by a first mass spectrometer is directed into a second mass spectrometer after being fragmented. Through the determination of the masses of the many fragments that can be produced by breaking one of the peptide bonds of the analyte peptide, the sequence can be determined.
The advantages of mass-spectrometry peptide sequencing are that blocked N-termini (a roadblock for Edman degradation) pose no problem, the rapid acquisition of sequence data, and characterization of common posttranslational modifications is possible.
A limitation of this method is that it is unable to distinguish between Ile and Leu (as they have identical residue masses), as well as difficulty in distinguishing Gln and Lys.
Sequence assembly: The "divide and conquer" strategy, piecing together overlapping peptide fragments produced by at least two different methods, is employed to deduce the complete sequence of the intact parent polypeptide. As a simple example, if treatment of a 10-residue peptide with trypsin yields fragments NYAN and ELFVHR, and treatment of the same peptide with chymotrypsin produces AN, ELF and VHRNY, then the intact peptide sequence must be ELFVHRNYAN.
Sequencing example: The first protein sequence, that of insulin, determined in the 1950s by Frederick Sanger.
Sequence databases
There are a number of important resources for online retrieval of information related to biological molecules, collectively indispensable, and which represent the product of bioinformatics. The National Center for Biotechnology Information (NCBI) and the Protein Data Bank (PDB) will serve as principal bioinformatic portals in this course. Other resources (as provided in VVP4e, Table 5-5 on p.115):
Protein Information Resource (PIR): http://pir.georgetown.edu/
UniProt: http://www.uniprot.org/
Sequence analysis: alignments
Pairwise comparison of sequences of proteins. Multiple sequence alignments and conserved residues.
Protein evolution
Evolutionary relationships are revealed by protein sequence comparisons. Phylogenetic trees can be constructed from multiple sequence alignments. Sequence comparisons provide information on protein structure and function.
Proteins evolve by duplication of genes or gene segments. Homologous proteins have protein sequences that have a high degree of identity and similarity (conservative substitutions). Not surprisingly, homology is indicative of evolutionary relationships. This is most evident in orthologous proteins, which are proteins that perform the same function in different organisms. For example, cytochrome c orthologs can be found in nearly every eukaryotic organism, and multiple sequence alignments of cytochrome c can be used to construct a phylogenetic tree - see VVP4e, Table 5-6 (pp.116-117) and Fig. 5-22 (p.119).
Among homologous proteins, paralogous proteins (paralogs) reside within the same organism. Paralogs arise as the result of gene duplication; hence they can evolve independently into divergent functional roles, in contrast to orthologous proteins
Proteins evolve at different rates, providing evolutionary clocks on vastly different time scales.
Domains, their duplication and divergence. A domain is a segment of a protein sequence that is conserved, apparently as a result of the evolutionary utility of the tertiary structure and its associated biological function. The smallest domains are the set by the typical minimum number of residues necessary to form a stable tertiary conformation (or 'fold"), usually cited as somewhere around 40 amino acids. The largest can be several hundred residues in length. Polypeptide chains of more than several hundred residues in length almost certainly fold into two or more domains. Our text at this point, at the end of Ch.5 (VVP4e, p.122), focused still on genetic information (or its translation into a protein sequence according to the genetic code), introduces domain shuffling as a evolutionary mechanism. Furthermore, domain shuffling may be a more rapid process by which protein diversity is generated than is gene duplication. Certainly domain duplication, followed by divergence, creates a greater multiplicity of independently evolving structural units. We'll revisit domains when we delve into protein tertiary structure in the next part of the course.