BIOCHEMISTRY TOPICS
Serine proteases
Nomenclature. The classes of proteases. Chymotrypsin, an example of the serine proteases. The cartalytic triad and its role in the serine protease mechanism. Specificity and convergent evolution in serine proteases.
Proteases are hydrolase enzymes that catalyze the hydrolysis of a peptide bond in a polypeptide or protein substrate. The peptide bond that is cleaved by a protease is called the scissile bond. The result of a protease-catalyzed hydrolysis of a peptide bond are two separate polypeptides, with new carboxyl and amino termini arising from bond scission.
The four classes of proteases
- Serine proteases (examples: chymotrypsin [EC 3.4.21.4], trypsin [EC 3.4.21.1], elastase; also subtilisin)
- Cysteine (or thiol) proteases (examples: papain [EC 3.4.22.2], mammalian cathepsins)
- Aspartyl proteases (a.k.a. carboxyl or acidic proteases) (examples: pepsin, renin, HIV protease)
- Metalloproteases (thermolysin, carboxypeptidase A, matrix metalloprotease [MMP])
Chymotrypsin: Covalent catalysis and the double displacement mechanism
The serine and cysteine proteases use a similar mechanism, and we'll treat chymotrypsin [EC 3.4.21.1] as a typical member of the serine proteases. Chymotrypsin follows a double-displacement (or "ping-pong") pattern, which is often observed for enzymes that make use of a covalent catalysis strategy.
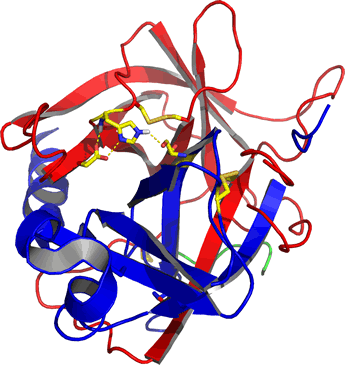
Right: Ribbon diagram of α-chymotrypsin, the mature, active form of the pancreatic enzyme. The residues of the catalytic triad (discussed below) and disulfide bonds are shown as sticks. The mature enzyme consists of three separate polypeptide chains that are the result of processing of a zymogen form of chymotrypsin called chymotrypsinogen. The A chain is colored green, the B chain (residues 16-146) is red, and the C chain (residues 149-245) is blue. The image was generated from the pdb entry 5cha.
The catalytic triad and its role in the serine protease mechanism
The catalytic triad provides a paradigm for the structural and chemical features of enzymes that allow them to facilitate a difficult reaction, peptide bond hydrolysis. The class of enzymes known as serine proteases - of which chymotrypsin is a member - have over the years been very thoroughly characterized by a wide variety of biochemical and structural methods. Residues of chymotrypsin important to its catalytic function were first identified using techniques of protein chemistry such as affinity labels in combination with a suitable assay for the enzyme's activity. Subsequently, the structures of chymotrypsin and other serine proteases revealed that the active sites of these enzymes shared a particular stereochemical arrangement of residues crucial to their activity.
This came to be known as the catalytic triad, as it consisted of the eponymous serine (Ser) residue, along with a histidine (His) and an aspartate (Asp) residue. A stepwise mechanism, described in detail on the catalytic triad webpage, was formulated that invokes special properties of the catalytic triad to explain the tremendous enhancement of the peptide hydrolysis reaction observed for the serine proteases.
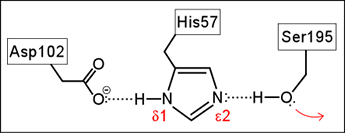
Left: Schematic diagram of the Ser-His-Asp catalytic triad of serine proteases. The red arrow indicates the nucleophilic property of the hydroxyl functional group of the serine residue.
Specificity in serine proteases
The preference of chymotrypsin for large hydrophobic residues at the P1 position of the substrate is accounted for by the existence of an appropriately located, properly sized non-polar cavity in the enzyme, serving as a specificity pocket, well-suited to accommodate the planar aromatic rings of Phe, Tyr, and Trp). In trypsin, this pocket has a negatively charged Asp residue at its bottom, explaining its specificity for Lys and Arg at P1. Elastase shows less well-defined specificity. It tends to prefer smaller, uncharged residues at P1. The elastase specificity pocket is constricted by Val and Thr residues, which replace Gly residues found in trypsin and chymotrypsin.
Convergent evolution in serine proteases
Comparison of the structures of different members of the class of serine proteases reveals close structural and functional similarities. The members within the trypsin family (including chymotrypsin, trypsin, and elastase) show sequence identities as high as 40%, and not surprisingly, are correspondingly similar in structure and functional properties. In a superposition of the main chain traces of chymotrypsin (pdb file 5cha.pdb) and elastase (pdb file 1est.pdb), the differences in positions of corresponding atoms in the best superposition of two structures is computed. For the Cα atoms of chymotrypsin and elastase, the root-mean-square difference (RMSD) value is 1.7 Å.
Interestingly, the catalytic triad seen in serine proteases has apparently arisen independently more than once in the course of evolution. The enzyme subtilisin, from the bacterium Bacillus subtilis, has a very similar Ser-His-Asp catalytic triad, despite being quite different in its overall structure. The residues that form the catalytic triad in subtilisin - Asp32, His64, and Ser221 - occur in a different order in the sequence, further evidence of its independent origin. The enzyme serine carboxypeptidase II from wheat germ has the same catalytic triad, and its structure is distinct from either trypsin or subtilisin. The triad in serine carboxypeptidase is composed of Ser146, Asp338, and His397.
Trypsin, chymotrypsin and other pancreatic proteases are secreted by the pancreas in an inactive precursor form called a zymogen. The zymogens provide examples of protein processing as a regulatory mechanism. The zymogen form is a proenzyme, with a "pro" form, similar to insulin, that is subject to proteolytic "editing". N-terminal and/or other fragments of the propolypeptide chain encoded by mRNA and produced by ribosomal synthesis are removed. In the case of trypsin, its zymogen form, called trypsinogen, is initially processed in the duodenum by enteropeptidase [EC 3.4.21.9], which removes a short N-terminal peptide from trypsinogen to yield active trypsin. Active trypsin is able to activate trypsinogen, as the processing reaction is an endopeptide cleavage following a lysine residue. The same is true for chymotrypsinogen, the zymogen form of chymotrypsin, except cleavage follows an arginine residue.