BIOCHEMISTRY TOPICS
The catalytic triad and its role in the serine protease mechanism
A detailed, stepwise account of the mechanism of serine proteases, describing and depicting the roles of each of the residues of the catalytic triad.
The catalytic triad provides a paradigm for the structural and chemical features of enzymes that allow them to facilitate a difficult reaction. The reaction in this case is hydrolysis of a peptide bond, which - although thermodynamically favorable - is kinetically inaccessible under normal physiological conditions. How do enzymes such as chymotrypsin manage to speed up this reaction by many, many orders of magnitude? The class of enzymes known as serine proteases - of which chymotrypsin is a member - have over the years been very thoroughly characterized by a wide variety of biochemical and structural methods. Residues of chymotrypsin important to its catalytic function were first identified using techniques of protein chemistry such as affinity labels in combination with a suitable assay for the enzyme's activity. Subsequently, the structures of chymotrypsin and other serine proteases revealed that the active sites of these enzymes shared a particular sterochemical arrangement of residues crucial to their activity. This came to be known as the catalytic triad, as it consisted of the eponyomous serine (Ser) residue, along with a histidine (His) and an aspartate (Asp) residue. A stepwise mechanism, described in detail on this page, was formulated that invokes special properties of the catalytic triad to explain the tremendous enhancement of the peptide hydrolysis reaction observed for the serine proteases.
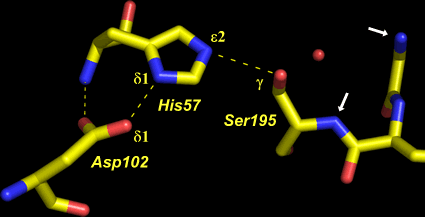
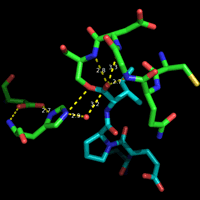
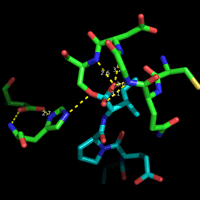
At left is shown the actual structure of the catalytic triad from the digestive enzyme chymotrypsin (based on the PDB entry 5cha.pdb). In this "sticks" representation of the structure, atoms are located at the vertices and ends of the sticks.
Carbon atoms are yellow, nitrogen blue, and oxygen red. The residues of the triad are labeled, as are several key atoms. The red sphere indicates the oxygen of a water molecule, which marks the approximate location of the acyl carbon of the scissile peptide bond when substrate binds. The white arrows point to amide nitrogens that form the so-called oxyanion hole. The water is located about 3.5 Å from the amide nitrogens and Oγ of Ser195.
The dashed lines represent hydrogen bonds. The distance between the heteroatoms Oδ1 of Asp102 and Nδ1 of His57 is 2.67 Å; the distance between Nε2 of His57 and Oγ of Ser195 is 3.02 Å. Note that there are no hydrogen atoms shown in the structure, which in this case was determined to 1.67 Å resolution. At this resolution, the hydrogens cannot be definitively located, so their positions must be inferred. This is an especially key point in this context, because in order to formulate a model for the mechanism, assumptions must be made about the locations of protons.
The next figure below is a schematic of the serine protease active site with a peptide substrate bound. Hydrogen atoms are included. The mechanism suggested by the catalytic triad has the following key features:
(i) His57 is unprotonated in the resting state of the enzyme.
(ii) His57 is poised to accept H+ from Ser195 as it attacks the carbonyl carbon of the scissile peptide bond.
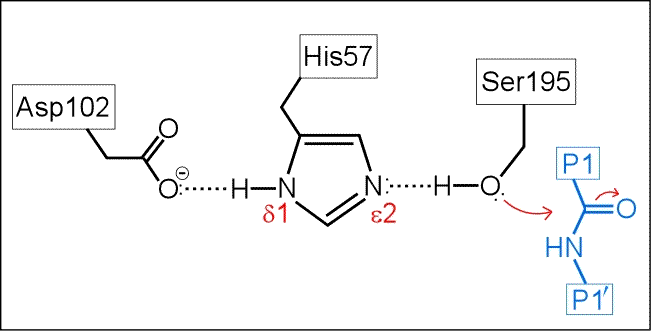
(iii) The role of Asp102 is three-fold: (i) it helps anchor His57 in its correct conformation, (ii) favors the tautomer of the neutral form of His57 with the proton on the δ1 N on the imidazole ring, and (iii) stabilization of the positive charge accumulating on His57 in the transition state as the Ser195 Oγ atom forms a covalent bond with the carbonyl carbon of the P1 residue of the substrate.
(iv) The "oxyanion hole" - two main-chain amide groups, one from Ser195 itself, the other from residue Gly193, stabilizes a presumed tetrahedral intermediate by favorable dipole-charge interactions. This could be manifested as two strong H-bonds.
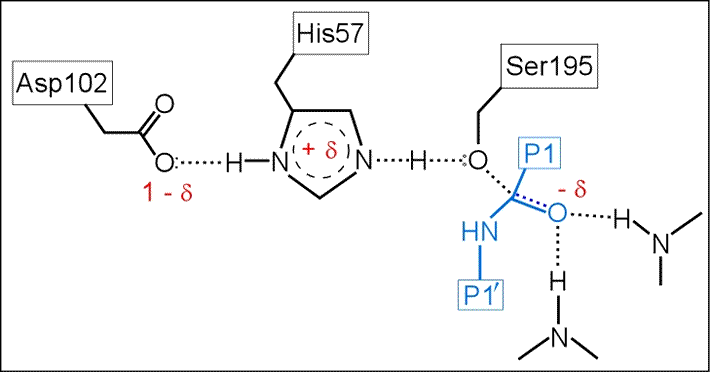
The above figure is a representation of the transition state between the initial attack of Ser195 on the carbonyl carbon of the substrate and the tetrahedral intermediate. The latter, illustrated in the figure below, is marked by the full development of a bond between the Oγ atom of Ser195 and the carbonyl carbon of the substrate. The catalytic triad functions as a charge relay system, as indicated by the distribution of partial charges.
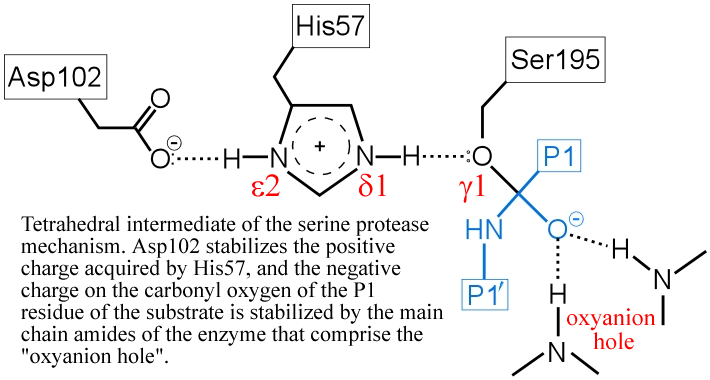
(v) The transient, protonated form of His57 is thought to transfer its proton to the amine leaving group
(vi) The form of the enzyme in which Ser195 is covalently attached to the N-terminal fragment of the cleaved substrate by an ester linkage is termed the acyl enzyme. Once the acyl enzyme is formed, and the C-terminal fragment of the substrate (first product) is released as depicted in (a), water (the second substrate) is bound in the active site. (b) A proton is transferred from water to His57, leading to the formation of the second tetrahedral intermediate.
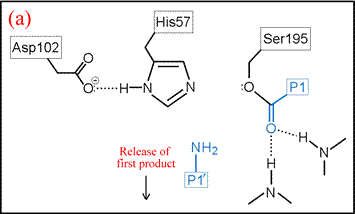
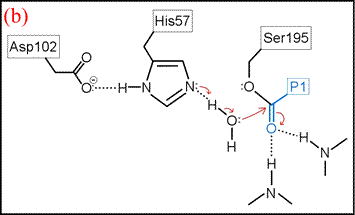
The deacylation is the reverse of acylation. A water molecule, taking the place of the released C-terminal fragment, attacks the carbonyl carbon of the acyl enzyme, again assisted by the histidine residue in its basic form.
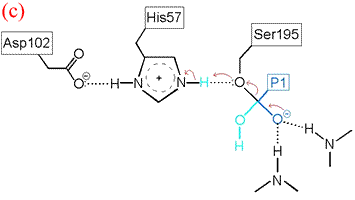
His57 then donates its extra proton to the γ oxygen of Ser195 as it leaves the substrate acyl carbon, forming the second product and completing the peptide bond hydrolysis. Upon release of the P1 fragment from the enzyme active site, a new peptide substrate can bind to the enzyme, and the hydrolytic mechanism can be reiterated.
Further explorations of enzyme mechanisms, regulation and kineticsThe relationship between structure and function is a constant theme in biochemistry, and nowhere is this more vivid than in the case of enzymes, whose structures give us a wealth of detailed information upon which proposed mechanistic models can be based. |
||||||
Serine proteases PyMOL scripts |
 |
 |
||||
|
 |
 |
||||