Lecture 33. Electron transport chain
Friday 2 December 2016
The mitochondrion. Electron carriers and electron transfer processes. The integral membrane complexes of the mitochondrial ETC. The exergonic process of the ETC drives the generation of a transmembrane hydrogen ion gradient.
Reading: VVP4e - Ch.18, pp.581-607.
Summary
In the next two lectures, we turn our attention to the bioenergetic payoffs that are the end result of oxidative metabolism. We begin here with a look at mitochondria and crucial role played by mitochondrial transporters, which move the necessary materials across the inner membrane. Then we undertake a survey of the electron transport chain, and (in the final lectures) ATP synthase and oxidative phosphorylation.
Mitochondria and mitochondrial transport
Mitochondria are organelles of eukaryotic cells that are energy specialists. The mitochondria is bounded by a relatively porous outer membrane. Inside the mitochondria is the highly folded and reticulated inner membrane.
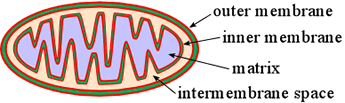
The intermembrane space is separated by the inner membrane from an inner compartment called the matrix. Specialized transporters - integral membrane proteins within the very protein-rich inner membrane - mediate the exchange of metabolites into and out of the matrix.
The inner membrane is otherwise highly impervious to the movement of ions or small molecules across it. Such compartmentalization has quite an important bearing on metabolism and its regulation in eukaryotic cells. For example, glycolysis takes place in the cytoplasm of cells. Pyruvate, the product of glycolysis, is transported into the mitochondrial matrix before it is oxidized by pyruvate dehydrogenase complex All of the citric acid cycle takes place in the matrix. If excess citrate accumulates in the matrix it can be exported to the cytosol to provide raw material for gluconeogenesis and fatty acid synthesis. Most of the ATP formed by oxidative phosphorylation, which takes place within the inner membrane and matrix, must be exported to the cytoplasm, while ADP and Pi must continually be imported to the matrix.
Taking a closer look at the inner mitochondrial membrane, we discuss its central role in the completion of the oxidation of metabolic fuel to water and carbon dioxide, which relies not only in its general impermeability and its embedded specific transport proteins, but also functions via a series of large integral membrane protein complexes that form a kind of "bucket brigade" that delivers electrons to oxygen. is in turn utilized to drive phosphorylation of ADP to produce ATP.
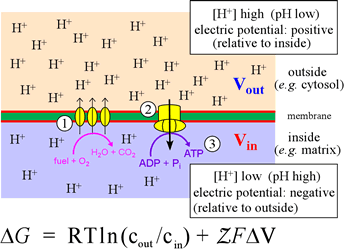
Right: The inner mitochondrial membrane divides the mitochondrial matrix (light blue, below membrane) from the intermembrane space (above, light pink). The catabolic pathways, converging on glycolysis and the citric acid cycle, convert food intake into CO2 and reduced electron carriers (mostly in the form of NADH, with some FADH2). The latter are the feedstock of electrons for the electron transport chain (ETC), represented in the figure by the three ovals (1). The ETC delivers the electrons from NADH and FADH2 ultimately to oxygen, an energy-yielding process that the complexes of the ETC couple to translocation of H+ from the matrix to the intermembrane space. This transmembrane pH difference, or electrochemical gradient, is itself a form of energy - its Gibbs free energy is given by the equation. The term ΔV is defined as ΔV = Vout − Vin. The definition of pH can be used to write the first term of the equation as −2.3026RTΔpH (at 25° C). This energy (2), in being dissipated through the proton channels of ATP synthase, drives the turning of its molecular generator that can form ATP molecules from ADP and Pi in the matrix (3).
The electron transport chain (ETC)
The ETC is responsible for the reduction of molecular oxygen by NADH This exergonic process is carried out in a precisely controlled, multistep manner that preserves much of the energy released in the form of a transmembrane electrochemical gradient. This feat is accomplished by four integral membrane protein complexes. Two mobile electron carriers shuttle electrons between these complexes, coenzyme Q (ubiquinone) and cytochrome c. Three of the four complexes translocate protons across the inner mitochondrial membrane, thus generating the transmembrane electrochemical gradient. Five types of prosthetic groups are involved in electron transfer within the complexes.
NADH-Q oxidoreductase (Complex I) [EC 1.6.5.3]
Official name: NADH dehydrogenase (ubiquinone). The largest complex >900 kD in size, with 46 distinct polypeptide chains. Cofactors: FMN, iron-sulfur (Fe-S) clusters. Protons pumped: about 4 H+ per pair of electrons transferred.
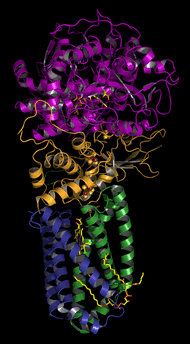
Succinate-Q reductase (Complex II)
140 kD, 4 subunits (pictured above). Cofactors: heme, quinone, iron-sulfur (Fe-S) clusters, and FAD. Does not pump H+.
Q-cytochrome c oxidoreductase (Complex III)
250 kD, 11 subunits. Cofactors: heme, Fe-S. Two H+ pumped per electron pair transferred. Also called cytochrome bc1 complex.
Cytochrome c oxidase (Complex IV)
160 kD, 13 subunits. Cofactors: hemes, copper ions. About 4 H+ are pumped out of the matrix per 2e− transferred.
Prosthetic groups of electron transfer
NADH, which feeds electrons at a relatively low biochemical standard reduction potential (E°′) into the ETC, is a pyridine nucleotide. The five types of prosthetic groups that participate in electron transfer in the respiratory chain are flavin nucleotides (FAD and FMN) quinones, iron-sulfur (Fe-S) clusters, heme and copper ion.
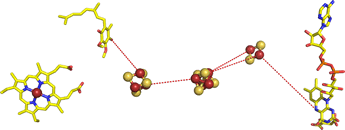
Right: The succinate-Q reductase (Complex II) structure, with the protein not displayed in order to clearly show its prosthetic groups and their arrangement. From left to right is heme a Q2 molecule (ubiquinone) with its nonpolar isoprenoid tail shortened to two five-carbon units), three iron-sulfur clusters, and FAD. The red dashes are meant to suggest electron transfer paths through the protein. The iron-sulfur clusters are separated by about 9 and 11 Å. The figure was drawn from pdb 1nek.